The unique transcription system of spirochaete bacteria may reveal new drug targets

Borreliosis, transmitted through ticks bites, becomes the talk in summertime. Antibiotic treatments for the disease are usually long, for example I had to take antibiotics for borreliosis for a month last summer. If the disease is not noticed in time, several long-term symptoms may remain. My unpleasant encounter with borreliosis is now driving me forward to study the special transcription system of spirochaete bacteria, which also includes the causative agent of borreliosis. In transcription, RNA polymerase (RNAP) builds RNA according to the DNA template, which then serves as instructions for different structures in cells. RNA can also act on other cellular events on their own. Transcription is the most regulated event of gene expression and this process is regulated by several different transcription factors (Belogurov and Artsimovitch 2015). A deeper understanding of the transcription system of spirochaetes could enable the development of new drugs.
The transcriptional system of spirochaete bacteria differs from well-known bacterial systems, but it has remained relatively unknown, despite its medical importance (Paster and Dewhirst 2000). In addition to borreliosis, spirochaetes also cause syphilis and leptospirosis. Since transcription is a very important and central event for cells, it cannot risk multiple mutations. This makes transcription a good target for drug development, and the RNAP of bacteria is recognized as a validated drug target (Ma et al. 2016). Since the transcription machines of the same type of bacteria resemble each other, I can use harmless Spirochaeta africana in my research, and the information obtained can be used to shed light on how the systems of dangerous spirochaetes function. Then I don’t have to stress about getting sick in my daily work and can just focus on science.
I study the spirochaete both at the level of transcriptional parts, where only the machinery that builds the RNA is involved, and at the level of whole cells. In transcription level experiments we add parts of the transcription system or known transcription inhibitors to a test tube and observe the course of the reaction. We have different ways to observe the reaction, one of them is based on fluorescence emitted by a fluorogen that binds to RNA that is built in the reaction (Huang et al. 2022). The RNA that is built is designed to have a structure that folds into a shape where the fluorogen can bind. We call this structure ‘’broccoli fluorescent light-up aptamer’’. The name comes from the green color of the fluorescence.
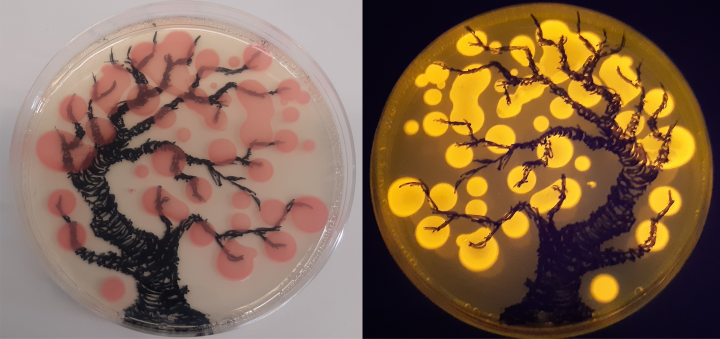
At the cellular level, we study the parts of the transcription system with the cell’s genes, their interaction, and the RNA produced by the cells. To study the parts of the transcription system we want to be able to transfer marked versions of its parts into the cells as genes. Then the cells would produce and use the marked proteins and we could get a better understanding of their role. We first tested genes with colorful marker proteins in well-known Escherichia coli bacteria and we saw that the marker protein showed up well both in normal room light as pink color and glowed brightly on a blue light table (pictured). Now we are trying to transfer the same gene to spirochaete bacteria by the means of an electric shock, so that we could see how the marker protein of the transferred gene is produced, after which we could similarly modify the parts of the transcription machinery and gain valuable insights of their role in the function of the cell.
Vilma Trapp
The author is a biochemistry doctoral student in Georgi Belogurov’s RNAP group at University of Turku. She studies the transcription system of spirochaete bacteria and its regulation in her thesis project.
References:
Belogurov, G.A. & Artsimovitch, I. (2015) Regulation of Transcript Elongation. Annu Rev Microbiol 69: 49–69.
Huang, Y.-H., Trapp, V., Puro, O., Mäkinen, J.J., Metsä-Ketelä, M., Wahl, M.C. & Belogurov, G.A. (2022) Fluorogenic RNA aptamers to probe transcription initiation and co-transcriptional RNA folding by multi subunit RNA polymerases. Meth Enzymol 675: 207-233
Ma, C., Yang, X. & Lewis, P.J. (2016) Bacterial Transcription as a Target for Antibacterial Drug Development. Microbiol Mol Biol Rev 80: 139–160.
Paster, B. & Dewhirst, F. (2000) Phylogenetic foundation of spirochetes. J Mol Microbiol Biotechnol 2: 341–4.